Summary
Almost 40 years ago, the first transgenic plant was generated through Agrobacterium tumefaciens-mediated transformation, which, until now, remains the method of choice for gene delivery in plants. Since then, optimized strains of Agrobacterium with additional (genetic) modifications have been developed whose main objective was to improve the efficiency of transformation, although there is also an optimized strain that reduces unwanted recombination of plasmids.
As a result, a collection of very useful strains has been created for transforming a wide variety of plant species, but it has also given rise to a confusing nomenclature of Agrobacterium strains. The latter is often misleading in choosing the most suitable variety for one’s transformation purposes. To overcome this problem, we provide a comprehensive overview of strain classification. We also indicate different strain modifications and their purposes, as well as the results obtained in terms of the long sensu transformation process.
In addition, we propose further improvements of the Agrobacterium-mediated transformation process and consider several valuable modifications, for example by circumventing a defence response in the plant. In this sense, we will discuss pattern-triggered immunity, pathogen-associated molecular pattern detection, hormone homeostasis and signalling, and reactive oxygen species in relation to Agrobacterium transformation. We will also explore alterations that increase agrobacterial transformation efficiency, reduce plasmid recombination, and improve biocontainment. Finally, we recommend the use of a modular system to better use the available knowledge for a successful transformation of the plant.
Keywords: Development of Agrobacterium strains; bacterial transformation; Plant transformation; Super-Agrobacterium; Stubborn transformation.
Materials and methods
- Bacterial strains
Rhizobia strains were isolated from root nodules of Phaseolus vulgaris grown in soil samples in a greenhouse according to standard procedures (Mhamdi, 1999). The agrobacterial strains (6a6, 10c2 and 22c1) were those used by Mhamdi (2005). These are nonnodulating agrobacteria that were isolated from common bean root nodules. The rhizobia inoculant consisted of a Rhizobium gallicum strain isolated from Bulla Regia (north-western Tunisia). The agrobacteria inoculant consisted of a mixture of strains 6a6, 10c2 and 22c2.
- Molecular characterization
Nineteen nodules were randomly recovered from each treatment (12 plants). Nodule bacteria were isolated and identified by 16S rRNA gene typing by PCR-restriction length polymorphism (RFLP) as previously described (Mhamdi, 2002). Symbiotic typing was achieved by PCR-RFLP of the nodC gene using MspI as previously described (Mhamdi, 2002).
- In vitro antibiosis assay
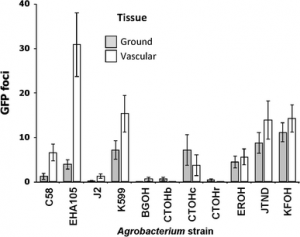
The plate overlay technique (Oresnik, 1999) was used to verify the in vitro inhibition of rhizobia by the three strains of agrobacteria (6a6, 10c2, 22c1). A saturated culture (48hold, OD630nm>1) of the indicator strain (rhizobia) grown in a liquid TY medium was diluted 10-3 in 5 ml of soft TY medium. The diluted suspension was poured onto a solid layer of TY in Petri dishes. Two hours later, 3 μL of a saturated culture of the producing strain (agrobacteria) was inoculated onto the soft agar surface. The zones of inhibition were measured after 4 days of incubation at 28°C. Each antibiosis test consisted of three repetitions with each strain of agrobacteria.
- Effectiveness test
Isolates Ma1.04, Ma1.06, Ma1.07, Ma1.12, Ma1.A05, Ma1.A10, Ma1.A32 and Ma1.A33 were evaluated for their symbiotic effectiveness in common beans. The experiment was carried out in 0.5 L plastic pots on sterile gravel as previously described (Mhamdi, 1999).
